

ExplORRNet: Explore miRNA in Breast and Gynecological Cancers
Welcome to ExplORRNet, a dedicated webtool to explore microRNAs (miRNAs) and their role in gynecological and breast cancers. MiRNAs are important regulators of gene expression, and understanding their dynamics is crucial for advancing cancer diagnosis and treatment.
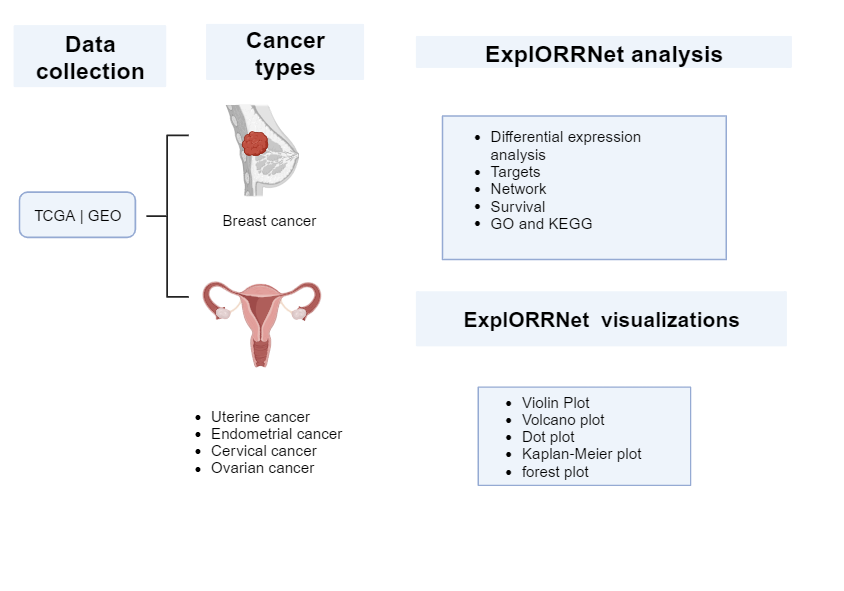
Figure 1 Graphical Abstract illustrating the schema of ExplORRNet
Our web tool is a comprehensive and user-friendly platform that seamlessly integrates transcriptomic and miRNA expression profiles from prominent databases including The Cancer Genome Atlas (TCGA) and the Gene Expression Omnibus (GEO).
A description of the data types and the number of patients used for the analysis is described in the table below:
Table 1: Sample number in each group for the five cancer types.
Why is this tool essential for cancer researchers and clinicians? Here's what ExplORRNet offers:
- To identify Dysregulated miRNAs: Identify miRNAs that are differentially expressed at various cancer stages, shedding light on their potential role in cancer development and progression.
- Explore Target Genes and Functions: Understand the biological functions affected by dysregulated miRNAs and their validated target genes, revealing vital molecular insights into gynecological and breast cancers.
- Patient Survival Analysis: Identify miRNAs associated with patient survival, aiding in the discovery of prognostic biomarkers for personalized treatment approaches.
- Network Analysis: Discover interconnected regulatory modules through miRNA-mRNA-lncRNA networks, unraveling the complex web of interactions influencing cancer biology.
- Non-Invasive Biomarkers: Delve into circulating miRNAs, offering potential non-invasive biomarkers for early cancer detection and monitoring.
- Breast Cancer Insights: Study breast cancer alongside gynecological cancers, leveraging similarities in hormonal regulation and risk factors, to gain a comprehensive view of women-specific cancers.
Whether you are a cancer biologist, bioinformatician, or clinician, ExplORRNet equips you with the tools to explore and interpret miRNA expression profiles in gynecological and breast cancers.
Visit https://mirna.cs.ut.ee to access ExplORRNet and unravel the miRNA landscape in gynecological and breast cancers.
If you find our tool useful, please cite our paper,
ExplORRNet: An interactive web tool to explore stage-wise miRNA expression profiles and their interactions with mRNA and lncRNA in human breast and gynecological cancers https://doi.org/10.1016/j.ncrna.2023.10.006
Do explore our recent paper on pan-cancer classification for deeper insights,
Tumor tissue-of-origin classification using miRNA-mRNA-lncRNA interaction networks and machine learning methods https://doi.org/10.3389/fbinf.2025.1571476
For any feedback or questions related to the tool please contact ankita.sunil.lawarde[*at]ut.ee
Tutorial Page
microRNA expression
Explore microRNA expression patterns of gynecological cancers and Breast cancer from The Cancer Genome Atlas (TCGA) project
Differentially regulated miRNA between Normal and all Tumor samples
Volcano Plot
Validated targets of differentially regulated miRNAs
Enrichment Analysis
Enrichment Plot
miRNA expression in Tissue
microRNA expression in clinical stage of tumor
Explore microRNA expression patterns of gynecological cancers and Breast cancer in different tumor stages from The Cancer Genome Atlas (TCGA) project
Differential regulation of miRNAs from TCGA data Subtype by tumor stage
Volcano Plot
Validated targets of differentially regulated miRNAs
Enrichment Analysis
Enrichment Plot
miRNA expression in Tissue
miRNA expression in serum, plasma, cell lysate and tumor tissue
Explore miRNA expression patterns of gynecological cancers and Breast cancer in serum, plasma, cell lysate and tumor tissue samples collected from GEO database
Differential regulation of miRNAs from GEO database
Volcano Plot
Validated targets of differentially regulated miRNAs
Enrichment Analysis
Enrichment Plot
miRNA expression in serum, plasma, cell lysate and tumor tissue
mRNA expression
Explore mRNA gene expression patterns of gynecological cancers and Breast cancer from The Cancer Genome Atlas (TCGA) project
Differentially regulated mRNA between Normal and all Tumor samples
Volcano Plot
mRNA expression in Tissue
mRNA expression in clinical stage of tumor
Explore mRNA gene expression patterns of gynecological cancers and Breast cancer in different tumor stages from The Cancer Genome Atlas (TCGA) project
Differential regulation of mRNAs from TCGA data Subtype by tumor stage
Volcano Plot
mRNA expression in Tissue
lncRNA expression
Explore lncRNA gene expression patterns of gynecological cancers and Breast cancer from The Cancer Genome Atlas (TCGA) project
Differentially regulated lncRNA between Normal and all Tumor samples
Volcano Plot
lncRNA expression in Tissue
lncRNA expression in clinical stage of tumor
Explore lncRNA gene expression patterns of gynecological cancers and Breast cancer in different tumor stages from The Cancer Genome Atlas (TCGA) project
Differential regulation of lncRNAs from TCGA data Subtype by tumor stage
Volcano Plot
lncRNA expression in Tissue
Survival Analysis
Survival probability and Cox-proportional model of miRNA expression in gynecological cancers and Breast cancer from The Cancer Genome Atlas (TCGA) project
Survival Plot
Survival Analysis
Survival probability and Cox-proportional model of mRNA expression in gynecological cancers and Breast cancer from The Cancer Genome Atlas (TCGA) project
Survival Plot
Survival Analysis
Survival probability and Cox-proportional model of lncRNA expression in gynecological cancers and Breast cancer from The Cancer Genome Atlas (TCGA) project
Survival Plot
Co-expression Network of miRNA-mRNA-lncRNA of Normal and tumor samples from breast and gynechological cancers.
Co-expression Network
Assortativity coefficient
Co-expression Network of miRNA-mRNA-lncRNA of Normal and all stages of tumor samples from breast cancer and Gynecological cancers.
Co-expression Network
Assortativity coefficient
Edge Table
Experimentally supported miRNAs and drug target associations in cancer collected from NoncoRNA database. For more information Please visit the NoncoRNA DB link.
Select Cancer Type
miRNA Drug Targtes
Protein Expression: Pathology data for Cancer-type and Tissue
Cancer Pathology Data from The Human Protein Atlas
Download data
Download miRNA, mRNA, lncRNA expression data, sample data. Be patient when downloading mRNA and lncRNA expression data.